SunlightCBM
@
|
 |
SunlightCBM is intended to be used inside a modern
unix environment, such as the GNU/Linux operating system.
Here we present examples using two simple networks, simple_network.xml
and simple_network_with_ext.xml, and some examples of
working with genome-scale models.
The SBML model in simple_network.xml describes the
following network :
name
|
reaction
|
explanation
|
EX_A
|
A ↔ |
exchange reaction for A
|
R1
|
2 A → B |
first internal reaction
|
R2
|
2 B → C |
second internal reaction
|
biomass
|
(0.2) B + (0.3) C → |
biomass demand reaction
|
Everything is happening in a single compartment, the cytosol (see later for a
generalisation). This is a very schematic model for anabolism.
Although you can view (and edit) simple_network.xml
in a text
editor, the
SunlightCBM perl scripts inspect.pl and export.pl
allow the contents of
the model to be easily extracted :
% perl inspect.pl
simple_network.xml
Simple network
(simple_network),
compartments = 1, metabolites = 3,
reactions = 4
In here '% ' represents the command line prompt.
More details can be retrieved using various options :
% perl
inspect.pl
simple_network.xml --compartments
[c]
Cytosol
% perl inspect.pl simple_network.xml
--metabolites
A_c
A
Cytosol
species-A_C123H938O11N7P2SR
B_c
B
Cytosol
species-B_C12H45OP2Zn
C_c
C
Cytosol
species-C_FeZn3Co
% perl inspect.pl simple_network.xml
--reactions
EX_A
exchange
reaction
for
A [c] : A <==>
R1
first
internal
reaction [c] : (2) A
--> B
R2
second
internal
reaction
[c] : (2) B --> C
biomass
biomass demand reaction
[c] : (0.2) B + (0.3) C -->
This first is a tab-delimited list of
the compartment abbreviations and full names. The second is a
tab-delimited list
comprising: the compartmentalised
metabolite abbreviation (this is where the compartment abbreviations
come in); the metabolite abbreviation without compartmentalisation; the
name of the compartment; and a descriptive name of the
metabolite.
The third is a tab-delimited list
comprising: the reaction abbreviation (this is used to identify
fluxes), a descriptive name for the reaction, and a schematic of the
actual reaction
following the format promoted by Palsson group.
You might notice the full metabolite names end with atomic
formulae. This is a convention adopted in some of the
SBML models from the Palsson group (in the present case the formulae
are hypothetical). The extra option
--with-formulae can be used to extract this information :
% perl inspect.pl simple_network.xml --metabolites
--with-formulae
A_c
A
Cytosol
species-A
C123H938O11N7P2SR
2688
1079
B_c
B
Cytosol
species-B
C12H45OP2Zn
205 58
C_c
C
Cytosol
species-C
FeZn3Co
0 0
Here the additional columns are the atomic formula, the atomic weight
(currently ignoring metal ions), and the number of atoms (also ignoring
metal ions).
The difference between inspect.pl and export.pl
is that the former can handle more than one SBML file at a time (try
it!), and the latter can export a model to cytoscape files taking
account of flux bounds (an example is shown below). Use the --help
option for more details.
Model optimisation
Now let's run a single optimisation (in fact the same one as used to
test the installation) :
% perl singleopt.pl
simple_network.xml "EX_A=(-10,--)"
6.25
This specifies that the SBML model is contained within simple_network.xml
and that the flux bounds for the exchange reaction EX_A should have a
minimum value -10, and an
unchanged maximum value (of ∞). By default, the script seeks out
the reaction name which matches the name 'biomass' and maximises the
flux through that reaction. It then returns this value of the
maximum flux, in this case 6.25.
Here it is again, using --verbose
to get more information :
% perl singleopt.pl simple_network.xml "EX_A=(-10,--)"
--verbose
SunlightCBM
(ParseSBML): Extracted
model Simple network
(simple_network) successfully
SunlightCBM
(ReadSBMLAndBounds):
from command line, EX_A = (-10,
Infinity)
SunlightCBM
(FindTarget):
Maximising flux through 'biomass demand
reaction' reaction (biomass)
SunlightCBM
(Initialise): LP
initialised for 3 x 4 problem
glp_simplex:
original LP has 3
rows, 4 columns, 7 non-zeros
glp_simplex:
presolved LP has 2
rows, 2 columns, 4 non-zeros
lpx_adv_basis:
size of triangular
part = 2
*
0:
objval
=
0.000000000e+00 infeas = 0.000000000e+00 (0)
*
1:
objval
=
6.250000000e+00 infeas = 0.000000000e+00 (0)
OPTIMAL
SOLUTION FOUND
SunlightCBM
(SolveProblem):
maximise biomass, solution was optimal,
result was 6.250000
6.25
Here it is again, this time with the actual fluxes :
% perl singleopt.pl simple_network.xml "EX_A=(-10,--)"
--fluxes
EX_A
-10
-0.625
non-basic,
at lower bound exchange
reaction for A
R1
5
0
basic
first internal
reaction
R2
1.875
0
basic
second internal
reaction
biomass
6.25
0
basic biomass demand reaction
The various columns are: flux (col 2), reduced
cost (col 3), status in LP terminology (col 4), and name (col
5). The reduced cost for a reaction gives the rate at which the
target
function (in this case the flux through the biomass demand reaction)
increases per unit flux increase through that reaction.
Reduced costs are discussed briefly in Palsson's book.
The tab-delimited output
can be captured and read into your favourite spreadsheet program, or
filtered for further analysis. For example, the output below is
piped into a gawk command, which splits each line into fields by tabs
(the -F "\t" option), then prints out the first two
fields ($1 and $2) as a string (%s),
followed
by
a
tab
(\t), followed by a numeric value (%f)
and
carriage return (\n) :
% perl singleopt.pl simple_network.xml "EX_A=(-10,--)"
--fluxes | gawk -F "\t" '{ printf "%s\t%f\n", $1, $2 }'
EX_A
-10.000000
R1
5.000000
R2
1.875000
biomass
6.250000
Finally, here are the shadow prices (one could use the option --with-formulae
here too) :
% perl singleopt.pl simple_network.xml "EX_A=(-10,--)"
--shadows
A_c
A
Cytosol
0.625
species-A-C123H938O11N7P2SR
B_c
B
Cytosol
1.25
species-B-C12H45OP2Zn
C_c
C
Cytosol
2.5
species-C-FeZn3Co
Note that the shadow
prices double from A to B to C, and the reduced cost for the A exchange
reaction (EX_A) is exactly minus the shadow price for A. This
is generally true, since reducing the flux through the
exchange reaction is the same as making the
associated metabolite more available. The shadow prices
can be determined also by direct solution of the dual problem :
% perl dualopt.pl simple_network.xml "EX_A=(-10,--)" -shadows
A_c
A
Cytosol
0.625
basic species-A-C123H938O11N7P2SR
B_c
B
Cytosol
1.25
basic species-B-C12H45OP2Zn
C_c
C
Cytosol
2.5
basic species-C-FeZn3Co
The result is the same as solving the primal (ie original)
problem. The new column gives the status of the dual variables in
LP terminology. Here is the dual solution again again, with some
more information
about what is happening :
% perl dualopt.pl simple_network.xml "EX_A=(-10,--)" --verbose
SunlightCBM
(ParseSBML): Extracted
model Simple network
(simple_network) successfully
SunlightCBM
(ReadSBMLAndBounds):
from command line, EX_A = (-10,
Infinity)
SunlightCBM
(FindTarget):
Maximising flux through 'biomass demand
reaction' reaction (biomass)
SunlightCBM
(InitialiseDual): LP
initialised for 4 x 3 problem
SunlightCBM
(SetDualBounds):
identified exchange reaction = EX_A
SunlightCBM
(SetDualBounds):
corresponding scale factor = 10
SunlightCBM
(SetDualBounds):
target = shadow price for A [Cytosol]
glp_simplex:
original LP has 4
rows, 3 columns, 7 non-zeros
glp_simplex:
presolved LP has 3
rows, 3 columns, 6 non-zeros
lpx_adv_basis:
size of triangular
part = 3
0:
objval
=
0.000000000e+00 infeas = 1.000000000e+00 (0)
2:
objval
=
6.250000000e-01 infeas = 0.000000000e+00 (0)
OPTIMAL
SOLUTION FOUND
SunlightCBM
(SolveProblem):
minimise A_c, solution was optimal, result
was 0.625000
dualopt.pl:
corresponding
maximised biomass flux was 6.25
0.625
Note that the direct solution of the dual problem is currently set up
to work only when there is a
single limiting exchange
reaction, since that can be used to identify a
unique metabolite shadow price as the target. Also, the final
output of the script is the shadow price for the target metabolite.
Now let's look at some examples where the LP problem does not have a
finite solution. Here's what happens if the growth rate is
unbounded, both without and with the --verbose option :
% perl singleopt.pl simple_network.xml "EX_A=(-Infinity,--)"
perl
singleopt.pl: flux unbounded,
when maximising biomass
Infinity
% perl singleopt.pl simple_network.xml "EX_A=(-Infinity,--)"
--verbose
SunlightCBM
(ParseSBML): Extracted
model Simple network
(simple_network) successfully
SunlightCBM
(ReadSBMLAndBounds):
from command line, EX_A = (-Infinity,
Infinity)
SunlightCBM
(FindTarget):
Maximising flux through 'biomass demand
reaction' reaction (biomass)
SunlightCBM
(Initialise): LP
initialised for 3 x 4 problem
glp_simplex:
original LP has 3
rows, 4 columns, 7 non-zeros
PROBLEM HAS NO
DUAL FEASIBLE
SOLUTION
SunlightCBM
(SolveProblem):
turning presolver off temporarily
*
0:
objval
=
0.000000000e+00 infeas = 0.000000000e+00 (3)
*
3:
objval
=
0.000000000e+00 infeas = 0.000000000e+00 (0)
PROBLEM HAS
UNBOUNDED SOLUTION
SunlightCBM
(SolveProblem):
maximise biomass
SunlightCBM
(SolveProblem):
solution was unbounded (GLP_UNBND), result
was Infinity
singleopt.pl:
flux unbounded,
when maximising biomass
Infinity
Here's what happens if the problem is infeasible, again without and
with the --verbose option :
% perl singleopt.pl simple_network.xml "EX_A=(10,--)"
perl
singleopt.pl: failed when
maximising biomass
Undefined
% perl singleopt.pl simple_network.xml "EX_A=(10,--)" --verbose
SunlightCBM
(ParseSBML): Extracted
model Simple network
(simple_network) successfully
SunlightCBM
(ReadSBMLAndBounds):
from command line, EX_A = (10,
Infinity)
SunlightCBM
(FindTarget):
Maximising flux through 'biomass demand
reaction' reaction (biomass)
SunlightCBM
(Initialise): LP
initialised for 3 x 4 problem
glp_simplex:
original LP has 3
rows, 4 columns, 7 non-zeros
PROBLEM HAS NO
PRIMAL FEASIBLE
SOLUTION
SunlightCBM
(SolveProblem):
turning presolver off temporarily
0:
objval
=
0.000000000e+00 infeas = 1.000000000e+00 (2)
PROBLEM HAS NO
FEASIBLE SOLUTION
SunlightCBM
(SolveProblem):
maximise biomass
SunlightCBM
(SolveProblem): no
feasible solution existed (GLP_NOFEAS)
SunlightCBM
(SolveProblem): no
primal feasible solution existed
(GLP_NOFEAS)
SunlightCBM
(SolveProblem):
problem was dual infeasible (GLP_INFEAS)
singleopt.pl:
failed when
maximising biomass
Undefined
Multicompartment models
The model in simple_network_with_ext.xml is similar to
that in simple_network.xml, shown above, except an extracellular
compartment has been added, a transporter reaction has been added
to allow A to move between the cytosol and the
extracellular space, and the exchange reaction now acts on
extracellular A :
name
|
reaction
|
explanation
|
EX_A
|
[e] : A ↔ |
exchange reaction for A
|
At
|
A [e] ↔ A [c]
|
transport reaction for A
|
R1
|
[c] : 2 A → B |
first internal reaction
|
R2
|
[c] : 2 B → C |
second internal reaction
|
biomass
|
[c] : (0.2) B + (0.3) C → |
biomass demand reaction
|
The optimal growth solution is basically the same as the previous model
:
% perl singleopt.pl simple_network_with_ext.xml
"EX_A=(-10,--)" --fluxes
At
10
0
basic
transport
reaction for A
EX_A
-10
-0.625
non-basic,
at lower bound exchange
reaction for A
R1
5
0
basic
first internal
reaction
R2
1.875
0
basic
second internal
reaction
biomass
6.25
0
basic biomass demand reaction
This model allows us to demonstrate some of the features of the
SunlightCBM scripts.
Adding and removing metabolites and reactions
The script mogrify.pl allows SBML models to be modified
by adding and
removing metabolites and reactions. We will see more extensive
use of this in the section discussing genome-scale models.
For the present time, let's add a metabolite (D) and a couple of
reactions (A[e] ↔ D[c] and [c] : (2) D → B) to the two-compartment
model above. Tab-delimited text files are used for listing the
metabolites or reactions to be added or removed. The format is
the same as the output from inspect.pl, although
obviously not all the
fields are used. Note: be careful if using emacs for editing
these files, as the tab characters are sometimes replaced by spaces,
which is not what you want! Here is a way of using the 'printf'
command for making a short files with strings (%s)
separated by tabs (%t) ending in a newline (%n)
:
% printf "%s\t%s\t%s\t%s\n" D_c D Cytosol "my new
metabolite" > metabs.dat
% printf "%s\t%s\t%s\n" At2 "alternate transport for A" "A[e]
<==> D[c]" > rxns.dat
% printf "%s\t%s\t%s\n" R3 "my new internal reaction" "[c] : (2)
D --> B" >> rxns.dat
% cat metabs.dat
D_c
D
Cytosol
my
new metabolite
% cat rxns.dat
At2
alternate
transport
for
A A[e] <==> D[c]
R3
my
new
internal
reaction [c] : (2) D --> B
Let's use mogrify.pl to add these to simple_network_with_ext.xml
and save the model for future use :
% perl mogrify.pl simple_network_with_ext.xml
--add-metabolites=metabs.dat --add-reactions=rxns.dat >
new_model.xml
Here is a single optimisation using the new model :
% perl singleopt.pl new_model.xml "EX_A=(-10,--)" --fluxes
At
10
0
basic
transport
reaction for A
At2
0
0
non-basic,
free
(unbounded) alternate transport for A
EX_A
-10
-0.625
non-basic,
at lower bound exchange
reaction for A
R1
5
0
basic
first internal
reaction
R2
1.875
0
basic
second internal
reaction
R3
0
0
basic
my new internal
reaction
biomass
6.25
0
basic biomass demand reaction
Obviously the added metabolite and reactions merely provide an
alternative pathway for the generation of B. This shows up for
instance if we stop the flux through the original transport reaction,
re-optimise, and compare the new flux distribution with the one above :
% perl singleopt.pl new_model.xml "EX_A=(-10,--)" At
--fluxes
At
10
0
basic
transport reaction for A
At2
0
0 non-basic, free
(unbounded) alternate transport for A
EX_A
-10 -0.625
non-basic, at lower bound exchange
reaction for A
R1
5
0 basic first internal
reaction
R2
1.875
0 basic second internal
reaction
R3
0
0 basic my new internal
reaction
biomass 6.25
0
basic biomass demand reaction
We exploit the shorthand notation where the name of a reaction by
itself as a parameter closes the reaction by setting the flux bounds to
(0, 0).
In fact At and At2 are a so-called 'synthetic lethal pair', as the
following sequence of single optimisations shows :
% perl singleopt.pl new_model.xml "EX_A=(-10,--)" At
6.25
% perl singleopt.pl new_model.xml "EX_A=(-10,--)" At2
6.25
% perl singleopt.pl new_model.xml "EX_A=(-10,--)" At At2
0
We can also see that there are alternate pathways (strictly speaking
alternate optima) in the new model by running a flux variability
study. Here the --prefac=1 option is used to force
the flux through the target biomass reaction to be the maximum possible
(ie 6.25) :
% perl variability.pl new_model.xml "EX_A=(-10,--)"
--prefac=1
At
0
10
bounded
transport reaction for A
At2
0
10 bounded alternate transport for A
EX_A
-10
-10 fixed exchange reaction for A
R1
0
5 bounded first internal reaction
R2
1.875 1.875
fixed second internal reaction
R3
0
5 bounded my new internal reaction
biomass 6.25
6.25 fixed
biomass demand reaction
The second and third columns give the lower and upper possible values
for the flux, the third column gives the status, and the last column
the descriptive name of the reaction.
Here, the two transporter reactions and the two alternative routes to
synthesis 'B' show up as fluxes with bounded variation reflecting the
alternate pathways (the other fluxes are fixed).
Deleting metabolites and reactions using mogrify.pl
follows a similar pattern to adding metabolites and reactions.
Some examples of this are given when genome-scale models are discussed.
One can regenerate an SBML model from the list of metabolites
and reactions generated by inspect.pl or export.pl
as the following
sequence shows :
% perl inspect.pl simple_network.xml --metabolites >
metabs.dat
% perl inspect.pl simple_network.xml --reactions > rxns.dat
% perl mogrify.pl --new-model --add-metabolites=metabs.dat
--add-reactions=rxns.dat | perl inspect.pl - --all
New Model
(new_model),
compartments = 1, metabolites = 3, reactions = 4
[c]
Cytosol
A_c
A
Cytosol
species-A-C123H938O11N7P2SR
B_c
B
Cytosol
species-B-C12H45OP2Zn
C_c
C
Cytosol
species-C-FeZn3Co
EX_A
exchange
reaction
for
A [c] : A <==>
R1
first
internal
reaction [c] : (2) A
--> B
R2
second
internal
reaction
[c] : (2) B --> C
biomass
biomass demand reaction
[c] : (0.2) B + (0.3) C -->
Note the use of a unix pipe to send the output of mogrify.pl
straight into inspect.pl. The '-'
option
makes
inspect.pl take its input from stdin.
Renaming compartments
It is possible to rename compartments using mogrify.pl,
both in terms of the full compartment name and the abbreviation.
In the convention promoted by the Palsson group, single characters
are used for compartment abbreviations (for example 'c' for cytosol,
and so on), and these are appended to the metabolite abbreviations with
a single underscore ("A_c" for example). If a model uses this
convention, we will say that it is in the standard format. SunlightCBM
extends this, by allowing multicharacter
compartment abbreviations. These are joined to the
metabolite abbreviation by a double underscore ("A__cyt" for
example). If a model uses multicharacter
compartment abbreviations, we will say that it is in the extended format.
Let's use mogrify.pl to change the abbreviation of the
cytosol compartment in simple_network.xml to "cyt",
converting the model from standard to extended format in the process :
% perl mogrify.pl simple_network.xml
--compartment="Cytosol->cyt" | perl inspect.pl - --all
Simple network
(simple_network__),
compartments = 1, metabolites = 3,
reactions = 4
[cyt]
Cytosol
A__cyt
A Cytosol
species-A-C123H938O11N7P2SR
B__cyt
B Cytosol
species-B-C12H45OP2Zn
C__cyt
C Cytosol
species-C-FeZn3Co
EX_A
exchange
reaction
for
A [cyt] : A <==>
R1
first
internal
reaction [cyt] : (2) A
--> B
R2
second
internal
reaction
[cyt] : (2) B -->
C
biomass
biomass demand reaction
[cyt] : (0.2) B + (0.3) C -->
Note that a double underscore is also added to the model id (ie
"simple_network__"). This is very important,
because SunlightCBM uses the presence of a double underscore in the
model abbreviation to indicate that a model is in the extended
format.
We can also rename the compartment keeping it in standard format, as
the following example shows :
% perl mogrify.pl
simple_network.xml
--compartment="Cytosol->(intracellular,i)" | perl inspect.pl - --all
Simple network
(simple_network),
compartments = 1, metabolites = 3,
reactions = 4
[i]
intracellular
A_i
A
intracellular
species-A-C123H938O11N7P2SR
B_i
B
intracellular
species-B-C12H45OP2Zn
C_i
C
intracellular
species-C-FeZn3Co
EX_A
exchange
reaction
for
A [i] : A <==>
R1
first
internal
reaction [i] : (2) A
--> B
R2
second
internal
reaction
[i] : (2) B --> C
biomass
biomass demand reaction
[i] : (0.2) B + (0.3) C -->
These models all perform exactly the same as the original one. By
the way, if you just want to convert a model into the extended format,
then use mogrify.pl with the --make-extended
option. Also, when making a new model the extended format
can be selected by using the --new-extended-model option.
Labelling
The extended format opens up the possibility of adding labels to
models, which can subsequently be joined
together in various ways. More examples are given later for
genome-scale models. Labelling is done with the --label
option of mogrify.pl, for example :
% perl mogrify.pl simple_network.xml --label=gut | perl
inspect.pl
- --all
Simple network
(simple_network__gut), compartments = 1, metabolites =
3, reactions = 4
[gut_c]
Cytosol__gut
A__gut_c
A
Cytosol__gut
species-A-C123H938O11N7P2SR
B__gut_c
B
Cytosol__gut
species-B-C12H45OP2Zn
C__gut_c
C
Cytosol__gut
species-C-FeZn3Co
EX_A__gut
exchange reaction for A [gut_c] : A
<==>
R1__gut
first internal reaction [gut_c]
: (2) A --> B
R2__gut
second internal
reaction [gut_c] : (2) B
--> C
biomass__gut
biomass
demand
reaction
[gut_c] : (0.2) B + (0.3) C
-->
Obviously many things have been changed in here : the compartment names
and abbreviations, the reaction abbreviations, and the model id;
uncompartmentalised metabolite abbreviations are not changed, rather the change is
made in the compartmentalised metabolite abbreviation. Although a
lot has changed, the model behaves exactly the same as before, as this
single optimisation shows (we also illustrate an occasionally useful
idiom whereby the --fluxes option is set and the output
is piped through grep -i biomass) :
% perl mogrify.pl simple_network.xml --label=gut | perl
singleopt.pl - "EX_A__gut=(-10,--)" --fluxes | grep -i biomass
biomass__gut
6.25
0
basic
biomass demand
reaction
Note that we have to specify "EX_A__gut" rather than "EX_A". The
labelling of exhange fluxes in this way can be avoided by using the --exclude-exchanges
option.
Further labels can be added, for example here we pipe the output of one
call to mogrify .pl into a second call, then inspect the
output :
% perl mogrify.pl simple_network.xml --label=gut | perl
mogrify.pl
- --label=human | perl inspect.pl - --all
Simple network
(simple_network__human_gut), compartments = 1,
metabolites = 3, reactions = 4
[human_gut_c]
Cytosol__human_gut
A__human_gut_c
A
Cytosol__human_gut
species-A-C123H938O11N7P2SR
B__human_gut_c
B
Cytosol__human_gut
species-B-C12H45OP2Zn
C__human_gut_c
C
Cytosol__human_gut
species-C-FeZn3Co
EX_A__human_gut
exchange
reaction
for
A [human_gut_c] : A <==>
R1__human_gut
first
internal
reaction
[human_gut_c] : (2) A
--> B
R2__human_gut
second
internal
reaction
[human_gut_c]
: (2)
B --> C
biomass__human_gut
biomass
demand
reaction
[human_gut_c] : (0.2) B + (0.3) C -->
The place where the label ends up can be changed with the --postfix
option. It is possible to combine the addition and removal of
metabolites and reactions with these compartment renaming and
labelling commands although care is advised since all combinations
have not been thoroughly tested. However, using compartment
renaming together with labelling is known to work sensibly, as the
examples using genome-scale networks show.
Genome-scale models
It has been a deliberate choice not to include genome-scale models with
SunlightCBM, since these models more properly belong to the groups who
have generated them. Here we illustrate the use of SunlightCBM
with the genome-scale models Ec_iJR904_GlcMM.xml for E. coli, and Sc_iND750_GlcMM.xml
for S. cerevisiae.
These are
available as Supplementary
Data
1 and Supplementary
Data
2 from the Nature
Protocols
article about the COBRA toolbox.
If
you
use these models in serious work,
you should certainly consider citing the following articles :
"An expanded genome-scale model of Escherichia coli K-12 (iJR904
GSM/GPR)", J. L. Reed, T. D. Vo, C. H. Schilling and B. Ø.
Palsson, Genome
Biology
4, R54.1 (2003).
"Reconstruction and validation
of Saccharomyces cerevisiae
iND750, a fully compartmentalized genome-scale metabolic model",
N. C. Duarte, M. J. Herrgard and B. Ø. Palsson, Genome
Research 14, 1298 (2004).
Other versions of these models, and models for other organisms, are
available from UCSD's BiGG database.
These
do
not
(typically?)
have biomass reactions in them. There
is also a minor but somewhat annoying name differences, for example
'mal-L' in the model exported from BiGG is 'mal_L' in the above SBML
models, and so on. In fact the whole problem of ontology is one which
remains to be resolved for these kind of models.
We suppose that you have got the E.
coli and S. cerevisiae
models somehow, and they are called Ec_iJR904_GlcMM.xml
and Sc_iND750_GlcMM.xml. Let's check these :
% perl inspect.pl Ec_iJR904_GlcMM.xml Sc_iND750_GlcMM.xml
Ec_iJR904_GlcMM.xml:
iJR904
(iJR904),
compartments
= 2, metabolites =
904, reactions = 1075
Sc_iND750_GlcMM.xml:
Sc_iND750
(Sc_iND750),
compartments
= 8,
metabolites = 1177, reactions = 1266
(this incidentally shows inspect.pl examining more than
one model at once). If you get something like this then you are
probably in good shape for the rest of this section.
These models cannot immediately be used by SunlightCBM. Firstly
the
exchange reactions are of the form A[e] ↔ A[b], where the
"b" is used to denote a boundary metabolite. Imposing the flux
balance condition on the boundary metabolites would force all the
exchange fluxes to zero. SunlightCBM does
not make a special exception for boundary metabolites, therefore
they must be removed from the models. Let's illustrate how this
can be done for iJR904 :
% perl inspect.pl Ec_iJR904_GlcMM.xml --metabolites |
gawk '$1 ~ /_b$/' > metabs.dat
% perl mogrify.pl Ec_iJR904_GlcMM.xml
--remove-metabolites=metabs.dat > new_model.xml
The first command identifies a list of all metabolites in Ec_iJR904_GlcMM.xml
whose compartmentalised abbreviations end with "_b"; this is done by
filtering the output of inspect.pl with a gawk
command. The second command removes these metabolites from Ec_iJR904_GlcMM.xml
to make new_model.xml. Comparing the old and new
models shows that 143 boundary metabolites have gone :
% perl inspect.pl Ec_iJR904_GlcMM.xml new_model.xml
Ec_iJR904_GlcMM.xml:
iJR904
(iJR904),
compartments
= 2, metabolites =
904, reactions = 1075
new_model.xml:
iJR904 (iJR904),
compartments = 2, metabolites = 761,
reactions = 1075
The boundary metabolites are silently dropped from the exchange
reactions, but will show
up if you run the above mogrify command with the --verbose
option.
The second reason why the models cannot be immediately used is that the
information they may contain about flux bounds is not used by
SunlightCBM. Rather the flux bounds in SunlightCBM default to
(−∞, ∞) for reversible reactions and (0, ∞) for irreversible reactions
(exchange reactions default to (0, ∞), unless the --open-exchanges
flag is used). To change these defaults, flux bounds have to be
specified separately either in the command line as we have already
seen or by reading in
tab-delimited text files. For example, by looking into Ec_iJR904_GlcMM.xml
(or otherwise) we
find that a number of exchange reactions should be fully open in order
for growth to occur. For convenience we have gathered these into
the following tab-delimited text file (some comment lines omitted) :
% cat open_exchanges.dat
. . .
EX_co2(e)
-Infinity
Infinity
EX_fe2(e)
-Infinity
Infinity
EX_h(e)
-Infinity Infinity
EX_h2o(e)
-Infinity
Infinity
EX_k(e)
-Infinity Infinity
EX_na1(e)
-Infinity
Infinity
EX_nh4(e)
-Infinity
Infinity
EX_pi(e)
-Infinity
Infinity
EX_so4(e)
-Infinity
Infinity
EX_o2(e)
-Infinity
Infinity
. . .
With these exchanges open, in only remains to specify a carbon/energy
source such as glucose which can be done on the command line. For
example, here is iJR904 optimised for growth with the above open
exchanges and with glucose uptake limited to 10 mmol / (gDW-hr) :
% perl singleopt.pl new_model.xml open_exchanges.dat
"EX_glc(e)=(-10,--)"
0.956973417061644
The result here means that the growth rate is 0.957 / hr, or a
doubling time of 60 × ln 2 / 0.957 = 43 minutes. Here it is
again with more output :
% perl singleopt.pl new_model.xml open_exchanges.dat
"EX_glc(e)=(-10,--)" --verbose
SunlightCBM
(ParseSBML): Extracted
model iJR904 (iJR904) successfully
SunlightCBM
(ReadBounds): from
open_exchanges.dat, EX_co2(e) =
(-Infinity, Infinity)
SunlightCBM
(ReadBounds): from
open_exchanges.dat, EX_fe2(e) =
(-Infinity, Infinity)
SunlightCBM
(ReadBounds): from
open_exchanges.dat, EX_h(e) =
(-Infinity, Infinity)
SunlightCBM
(ReadBounds): from
open_exchanges.dat, EX_h2o(e) =
(-Infinity, Infinity)
SunlightCBM
(ReadBounds): from
open_exchanges.dat, EX_k(e) =
(-Infinity, Infinity)
SunlightCBM
(ReadBounds): from
open_exchanges.dat, EX_na1(e) =
(-Infinity, Infinity)
SunlightCBM
(ReadBounds): from
open_exchanges.dat, EX_nh4(e) =
(-Infinity, Infinity)
SunlightCBM
(ReadBounds): from
open_exchanges.dat, EX_pi(e) =
(-Infinity, Infinity)
SunlightCBM
(ReadBounds): from
open_exchanges.dat, EX_so4(e) =
(-Infinity, Infinity)
SunlightCBM
(ReadBounds): from
open_exchanges.dat, EX_o2(e) =
(-Infinity, Infinity)
SunlightCBM
(ReadSBMLAndBounds):
from command line, EX_glc(e) = (-10,
Infinity)
SunlightCBM
(FindTarget):
Maximising flux through 'BiomassEcoli'
reaction (BiomassEcoli)
SunlightCBM
(Initialise): LP
initialised for 761 x 1075 problem
glp_simplex:
original LP has 761
rows, 1075 columns, 4503 non-zeros
glp_simplex:
presolved LP has 487
rows, 706 columns, 2861 non-zeros
lpx_adv_basis:
size of triangular
part = 462
*
0:
objval
=
0.000000000e+00 infeas = 0.000000000e+00 (25)
*
159:
objval =
9.569734171e-01 infeas = 6.369887142e-10 (8)
OPTIMAL
SOLUTION FOUND
SunlightCBM
(SolveProblem):
maximise BiomassEcoli, solution was
optimal, result was 0.956973
0.956973417061644
Two recent publications suggest
simple updates to the original iJR904 model :
"Systematic assignment of thermodynamic constraints in metabolic
network models", A. Kümmel,
S. Panke and M. Heinemann, BMC
Bioinformatics 7, 512 (2006).
The
authors
suggest
the
reaction 'GALU' is irreversible.
"Genome-scale in silico models of E.
coli have multiple equivalent phenotypic states: assessment of
correlated reaction subsets that comprise network states", J. L. Reed
and B. Ø. Palsson, Genome
Research 14, 1797 (2004).
The
authors
suggest
a
number of reactions that can be removed to
eliminate so-called type III cycles.
We have provided some tab-delimited text files to make it
easier to update the iJR904 model along these lines (some comment lines
omitted) :
% cat iJR904_rxn_add.dat
. . .
GALU
UTP-glucose-1-phosphate
uridylyltransferase
[c]
: g1p + h + utp -->
ppi + udpg
. . .
% cat iJR904_rxn_remove.dat
. . .
ABUTt2
4-aminobutyrate
transport in via proton symport
ACCOAL
acetate-CoA ligase
(ADP-forming)
ADK1
adenylate
kinase
ADNt2
adenosine
transport
in
via proton symport
ALARi
alanine
racemase
(irreversible)
CYTDt2
cytidine transport in
via proton symport
GALUi
UTP-glucose-1-phosphate
uridylyltransferase
(irreversible)
GLUt4
Na+/glutamate
symport
INSt2
inosine
transport
in
via proton symport
LCADi
lactaldehyde
dehydrogenase
PROt4
Na+/Proline-L
symporter
SERt4
L-serine
via
sodium
symport
THMDt2
thymidine transport
in via proton symport
THRt4
L-threonine
via
sodium
symport
URAt2
uracil
transport
in
via proton symport
URIt2
uridine
transport
in
via proton symport
VPAMT
Valine-pyruvate
aminotransferase
. . .
(In iJR904_scale_factors.dat we also provide a list of
the metabolites
in iJR904 that represent multiple copies of the actual
molecules.)
These files can be used to make an up-to-date iJR904 model as follows :
% perl mogrify.pl new_model.xml
--add-reactions=iJR904_rxn_add.dat
--remove-reactions=iJR904_rxn_remove.dat > iJR904.xml
If you use the updated iJR904 model in serious work, you should
obviously cite the above two
publications
in addition to the original one.
For
convenience
the
steps
to
go from Ec_iJR904_GlcMM.xml
to an updated iJR904 model which works with SunlightCBM have been built
into the Makefile. Thus 'make ec' will make the
above file iJR904.xml from Ec_iJR904_GlcMM.xml
(using
a temporary xml file instead of new_model.xml). The
following shows this in action (assuming iJR904.xml
has
not yet been generated):
% make ec
perl
inspect.pl
Ec_iJR904_GlcMM.xml --metabolites | gawk '$1 ~
/_b$/' > metabs.dat
perl
mogrify.pl
Ec_iJR904_GlcMM.xml --remove-metabolites=metabs.dat
>
temp.xml
perl
mogrify.pl temp.xml
--add-reactions=iJR904_rxn_add.dat
--remove-reactions=iJR904_rxn_remove.dat > iJR904.xml
rm -f
metabs.dat temp.xml
% perl singleopt.pl iJR904.xml open_exchanges.dat
"EX_glc(e)=(-10,--)"
0.956973417043535
% perl singleopt.pl iJR904.xml open_exchanges.dat
"EX_glc(e)=(-10,--)" ">EX_o2(e)"
0.298843355048565
% perl singleopt.pl iJR904.xml open_exchanges.dat
"EX_glc(e)=(-10,--)" "ATPM=(7.6,--)"
0.921948095050457
In these, the first of the optimisations is the one done earlier.
In the second one, growth under anaerobic conditions is simulated -
note
that the O2 exchange flux overwrites the one
in open-exchanges.dat because it occurs later in the
command line, and we use another of the shorthand notations to set the
flux bounds to (0, ∞). In the third one, the minimum flux
through the non-growth-associated maintenance (NGAM) reaction is forced
to be 7.6 mmol / (gDW-hr). If
you are wondering what the reaction ATPM is, then it is easy to find
out :
% perl inspect.pl iJR904.xml --reactions |
grep ATPM
ATPM
ATP_maintenance_requirement
[c]
: atp + h2o
--> adp + h + pi
A similar approach can be taken for S.
cerevisiae, except that we don't have any reaction
updates. Again for convenience the steps to make a working model
have been gathered into the Makefile, as the following sequence shows
(this assumes you have Sc_iND750_GlcMM.xml and iND750.xml is
not yet generated) :
% make sc
perl
inspect.pl
Sc_iND750_GlcMM.xml --metabolites | gawk '$1 ~
/_b$/' > metabs.dat
perl
mogrify.pl
Sc_iND750_GlcMM.xml --remove-metabolites=metabs.dat
>
iND750.xml
rm -f
metabs.dat
% perl singleopt.pl
iND750.xml open_exchanges.dat
"EX_glc(e)=(-10,--)"
SunlightCBM
(ReadBounds): in
open_exchanges.dat, didn't find reaction
EX_fe2(e) to reset bounds
0.973233759037675
It turns out that the same open_exchanges.dat can be used
with iND750 as with iJR904 since the exchange reaction abbreviations
are all the same. As you can see, the only issue is that there is
no Fe2+ exchange reaction in iND750.
This results in a warning
message but is
otherwise ignored. Again, if you use the iND750 model for serious
work, you
should cite the original publication.
Advanced topic - joining two models
Using labelling it is relatively easy to join two models
together. The trick is to let the models share a common
extracellular
compartment. Here, in outline, is one possible approach :
- Label both models, keeping the extracellular compartments
un-relabelled.
- Export the reactions and metabolites from both models, and
concatenate them.
- Modify the biomass reactions, and add a new joint biomass
reaction.
- Generate a new model from the metabolite and reaction lists.
Here we show how this works in practice, by joining the E. coli and S. cerevisiae models. The
following presumes that you have generated iJR904.xml
and iND750.xml as discussed in the previous
section.
The following two sequences show the first step, making and testing
labelled versions of the models :
% perl mogrify.pl iJR904.xml --label=ecoli
--compartment="Extra_organism->(Extra_organism, ext)"
--exclude-exchanges > model1.xml
% perl inspect.pl model1.xml --compartments
[ecoli_c]
Cytosol__ecoli
[ext]
Extra_organism
% perl singleopt.pl model1.xml open_exchanges.dat
"EX_glc(e)=(-10,--)"
0.956973417043535
% perl mogrify.pl iND750.xml --label=yeast
--compartment="Extra_organism->(Extra_organism, ext)"
--exclude-exchanges > model2.xml
% perl inspect.pl model2.xml --compartments
[ext]
Extra_organism
[yeast_c]
Cytosol__yeast
[yeast_g]
Golgi_Apparatus__yeast
[yeast_m]
Mitochondria__yeast
[yeast_n]
Nucleus__yeast
[yeast_r]
Endoplasmic_Reticulum__yeast
[yeast_v]
Vacuole__yeast
[yeast_x]
Peroxisome__yeast
% perl singleopt.pl model2.xml open_exchanges.dat
"EX_glc(e)=(-10,--)"
SunlightCBM
(ReadBounds): in
open_exchanges.dat, didn't find reaction
EX_fe2(e) to reset bounds
0.973233759037675
Now we extract the metabolites and reactions from both models and
concatenate them into single files :
% perl inspect.pl model1.xml --metabolites >
metabs.dat
% perl inspect.pl model1.xml --reactions > rxns.dat
% perl inspect.pl model2.xml --metabolites >>
metabs.dat
% perl inspect.pl model2.xml --reactions >>
rxns.dat
At this point we can make a joint model :
% perl mogrify.pl --new-extended-model
--add-metabolites=metabs.dat --add-reactions=rxns.dat >
new_model.xml
% perl inspect.pl new_model.xml
New Model
(new_model__),
compartments = 9, metabolites = 1737,
reactions = 2239
We can grow each individual model.
% perl singleopt.pl new_model.xml open_exchanges.dat
"EX_glc(e)=(-10,--)" --target=/BiomassEcoli/ --fluxes | grep -i biomass
BiomassEcoli__ecoli
0.995789136591818
0
basic
BiomassEcoli
biomass_SC4_bal__yeast
0
-0.801102593636717
non-basic,
at lower
bound biomass_SC4_bal
% perl singleopt.pl new_model.xml open_exchanges.dat
"EX_glc(e)=(-10,--)" --target=/biomass_SC4/ --fluxes | grep -i biomass
BiomassEcoli__ecoli
0
-0.889614157459108
non-basic,
at lower
bound BiomassEcoli
biomass_SC4_bal__yeast
1.08057548905981
0
basic
biomass_SC4_bal
Interestingly the growth rates are
faster in the joint models than they are in the separate models.
The reason is that each organism can use pathways in the other
organism. This can be proved by re-running the models but
shutting down the appropriate cytosol-extracellular transporter
reactions, in which
case the original growth rates are recovered. We show how this
works for the growth of yeast. First we extract the relevant
transport reactions and make a tab-delimited file suitable to shut them
down :
% perl inspect.pl new_model.xml --reactions | gawk -F "\t"
'$3 ~ /\[ecoli_c\]/ && $3 ~ /\[ext\]/ { printf "%s\t0\t0\n", $1
}' > shutdown_transports.dat
The components of the gawk
command here are: an option to break into fields on tabs (-F "\t");
a
rule
to
select
lines whose reaction schematic (3rd field) contains
both [ecoli_c] and [ext] ($3 ~ /\[ecoli_c\]/ && $3 ~
/\[ext\]/);
and an action which prints out the reaction abbreviation (first field)
followed by
two tab separated zeros as a suitable format for resetting the flux
bounds (printf "%s\t0\t0\n", $1). The result is
saved in shutdown_transports.dat.
This contains 192 transporter reactions :
% wc -l shutdown_transports.dat
192
shutdown_transports.dat
% head -5 shutdown_transports.dat
12PPDt__ecoli
0
0
ACACt2__ecoli
0
0
ACALDt__ecoli
0
0
ACGApts__ecoli
0
0
ACMANApts__ecoli
0
0
Now we recalculate the maximum yeast growth rate including shutdown_transports.dat
:
% perl singleopt.pl new_model.xml
open_exchanges.dat "EX_glc(e)=(-10,--)" shutdown_transports.dat
--target=/biomass_SC4/ --fluxes | grep -i biomass
BiomassEcoli__ecoli
0
0
non-basic,
at lower
bound BiomassEcoli
biomass_SC4_bal__yeast
0.973233759037701
0
basic
biomass_SC4_bal
The
result is the same as the yeast
model on its own. This demonstrates that the E. coli
transporter reactions are essential
to obtain the higher growth rate of yeast in the joint model. A
similar exercise can be done for the growth of E.
coli.
Of course what we really want to do is grow the organisms jointly by
combining the biomass reactions. A trick to do this is to
introduce a fake 'biomass' metabolite that is generated by the
individual biomass reactions, and consumed by a new joint biomass
reaction. Let's see how to do this, working with the intermediate
tab-delimited metabolite and reaction files of the above model.
Using your favourite text editor or otherwise, add the following two
lines to metabs.dat :
biomass__ecoli_c biomass
Cytosol__ecoli E. coli biomass
biomass__yeast_c biomass
Cytosol__yeast S. cerevisiae biomass
If you use cut & paste, please note there are four tab-separated
fields here, the last field being a descriptive name. These lines
add two instances of the generic metabolite 'biomass', one for each of
the cytosol compartments. Now, edit the biomass reactions in rxns.dat
so that ' + biomass' is added to the right hand sides
(note that these reactions are compartmentalised to the individual
cytsols), and add a new joint biomass reaction to rxns.dat
to consume these :
joint_biomass Joint biomass (0.2)
biomass[ecoli_c] + (0.8) biomass[yeast_c] -->
Again, if you use cut & paste, note there are 3 tab-separated
fields, with the second field being a descriptive name (in this case
"Joint biomass") and the third field being the reaction. The
stoichiometry coefficients in here are x
=
gDW (E. coli) / gDW (total),
and 1 − x = gDW (S. cerevisiae) / gDW (total).
We have chosen x = 0.2
arbitrarily here
to
illustrate the approach.
Now we can build the model again, and this time optimise for flux
through the new joint reaction :
% perl mogrify.pl --new-extended-model
--add-metabolites=metabs.dat --add-reactions=rxns.dat > new_model.xml
% perl singleopt.pl new_model.xml open_exchanges.dat
"EX_glc(e)=(-10,--)" --target=joint_biomass --fluxes | grep -i biomass
BiomassEcoli__ecoli
0.21767414605297
0
basic
BiomassEcoli
biomass_SC4_bal__yeast
0.87069658421188
0
basic
biomass_SC4_bal
joint_biomass
1.08837073026485
0
basic
Joint biomass
We see that the fluxes is going through the individual biomass
reactions are, respectively, 20% and 80% of the flux
through the joint reaction.
In case you are wondering, the growth
rate is the flux through the
joint biomass reaction. The calculation can be repeated
as a function of x and the
result is shown in the figure below (the perl
script scan.pl is set up to do this calculation, but is
not part of
SunlightCBM as such). There is a weak maximum growth rate around x = 0.35.
For convenience, the steps to make this joint model both
with and without the joint biomass reaction are included in the
Makefile, and can be run by typing 'make ec_plus_sc'
(the editing of the metabolite and reaction files is done using printf
and sed commands; x = 0.12345
is set in the Makefile but can be changed).
Advanced topic - putting one model inside another one
In a similar way to joining models, it is possible to put one model
inside another one. This can be done by making the extracellular
compartment of one model the same as the cytosol of the second
model. Here, in outline, is one possible approach to do this :
- Label the 'inner' model making sure the extracellular compartment
is renamed appropriately.
- Export the metabolites and reactions, and remove the exchange
reactions.
- Import the metabolites and reactions into the 'outer' model.
- Modify the biomass reactions as appropriate.
We illustrate this by putting E.
coli inside S.
cerevisiae. Again, the
following presumes that you have generated iJR904.xml
and iND750.xml models as discussed previously.
Firstly we make a suitably renamed version of iJR904.xml.
% perl mogrify.pl iJR904.xml --label=ecoli
--compartment="Extra_organism->(Cytosol, c)" --exclude-exchanges
> model1.xml
% perl inspect.pl model1.xml --compartments
[c]
Cytosol
[ecoli_c]
Cytosol__ecoli
% perl singleopt.pl model1.xml open_exchanges.dat
"EX_glc(e)=(-10,--)" --fluxes | grep -i biomass
BiomassEcoli__ecoli
0.956973417043535
0
basic
BiomassEcoli
Now we export the metabolites and reactions (the gawk command in here
has the effect of removing the exchange reactions) :
% perl inspect.pl model1.xml --metabolites >
metabs.dat
% perl inspect.pl model1.xml --reactions | gawk '$1 !~
/EX_/' > rxns.dat
If we try to import these into iND750.xml, we run into
trouble because iND750.xml uses the standard format and
does not allow multicharacter compartment abbreviations.
Therefore we should first promote iND750.xml to use the
extended format, and check it, as shown here :
% perl mogrify.pl --make-extended iND750.xml > model2.xml
% perl singleopt.pl model2.xml open_exchanges.dat
"EX_glc(e)=(-10,--)" --fluxes | grep -i biomass
SunlightCBM
(ReadBounds): in
open_exchanges.dat, didn't find reaction
EX_fe2(e) to reset bounds
biomass_SC4_bal
0.973233759037675
0
basic
biomass_SC4_bal
Now we can do the import :
% perl mogrify.pl model2.xml
--add-metabolites=metabs.dat --add-reactions=rxns.dat >
new_model.xml
% perl inspect.pl new_model.xml
Sc_iND750
(Sc_iND750__),
compartments = 9, metabolites = 1727,
reactions = 2181
Again, we can grow both organisms :
% perl singleopt.pl new_model.xml open_exchanges.dat
"EX_glc(e)=(-10,--)" --target=/BiomassEcoli/ --fluxes | grep -i biomass
SunlightCBM
(ReadBounds): in
open_exchanges.dat, didn't find reaction
EX_fe2(e) to reset bounds
BiomassEcoli__ecoli
1.37441367226407
0
basic
BiomassEcoli
biomass_SC4_bal
0
-0.809171045529626
non-basic,
at lower
bound biomass_SC4_bal
% perl singleopt.pl new_model.xml open_exchanges.dat
"EX_glc(e)=(-10,--)" --target=/biomass_SC4/ --fluxes | grep -i biomass
SunlightCBM
(ReadBounds): in
open_exchanges.dat, didn't find reaction
EX_fe2(e) to reset bounds
BiomassEcoli__ecoli
0
-1.12345672193585
non-basic,
at
lower bound BiomassEcoli
biomass_SC4_bal
1.63247226769719
0
basic
biomass_SC4_bal
In these cases the growth rate of the two components is significantly
enhanced over the individual growth rates (but see below), again
because
one organism can use the pathways in the other organism. We can
verify this for the growth of yeast by shutting down the E. coli transporter
reactions, as above. It doesn't make sense to check the other way
though
since the yeast transporter reactions are the only way that material
can enter the yeast cytosol, and from there the E. coli cytosol. At this
point you might question the lack
of an Fe2+ transporter (and possibly other
transporters) in the yeast model.
However,
it
turns
out
this E. coli
model (at least) does not need these 'missing' transporter reactions to
grow;
if it did then no growth possible.
For the last step we can proceed in a very similar way to the previous
section and add fake 'biomass' metabolites to keep track of
the two biomass reactions, and a new joint biomass reaction to consume
these. Using your favourite text editor or otherwise, add the
following two
lines to metabs.dat :
biomass__ecoli_c biomass
Cytosol__ecoli E. coli biomass
biomass__c
biomass
Cytosol S.
cerevisiae biomass
These lines
add two instances of the generic metabolite 'biomass', one for each of
the cytosol compartments. Now, let's export the yeast biomass
reaction to rxns.dat,
% perl inspect.pl iND750__ext.xml --reactions | grep -i
biomass >> rxns.dat
and edit the two biomass reactions in rxns.dat
so that ' + biomass' is added to the right hand sides
(note that these reactions are compartmentalised to the individual
cytosols). Also, add a new joint biomass reaction to consume
these
:
joint_biomass Joint biomass (0.1)
biomass[ecoli_c] + (0.9) biomass[c] -->
The stoichiometry coefficients in here are
again mass fractions, with x
=
gDW (E. coli) / gDW (total),
and 1 − x = gDW (S. cerevisiae) / gDW (total).
We have chosen x = 0.1
arbitrarily here
to
illustrate the approach.
If we rebuild the model and re-optimise we find :
% perl mogrify.pl model2.xml
--add-metabolites=metabs.dat --add-reactions=rxns.dat > new_model.xml
% perl singleopt.pl new_model.xml open_exchanges.dat
"EX_glc(e)=(-10,--)" --target="joint_biomass" --fluxes | grep -i biomass
SunlightCBM
(ReadBounds): in
open_exchanges.dat, didn't find reaction
EX_fe2(e) to reset bounds
BiomassEcoli__ecoli
0.160238602115474
0
basic
BiomassEcoli
biomass_SC4_bal
1.44214741903927
0
basic
biomass_SC4_bal
joint_biomass
1.60238602115474
0
basic
Joint biomass
We see that the growth rate is apparently considerably faster than
yeast on its
own. The growth rate can be calculated as a function of the mass
fraction, and the result is shown in the figure
below, compared to the case of the previous section (the perl
script scan.pl is set up to do this calculation, but is
not part of
SunlightCBM as such).
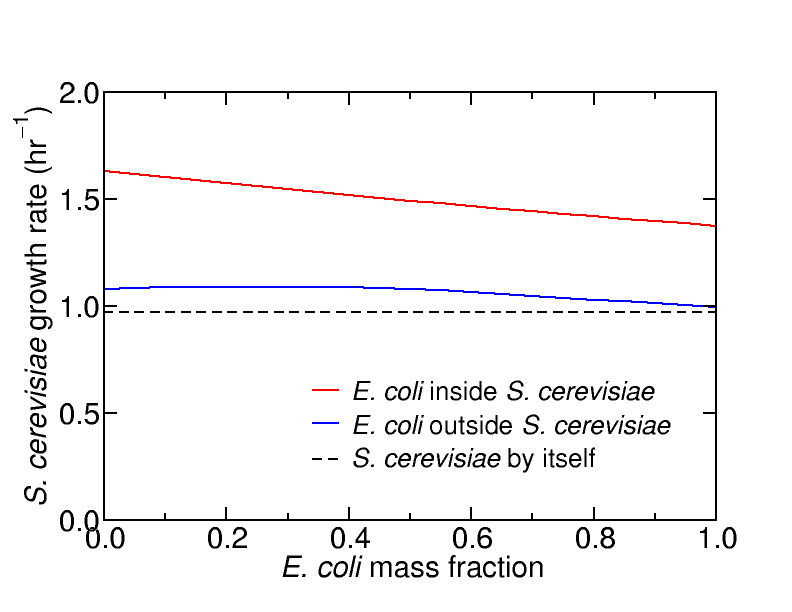
Growth
rates of joined E. coli and S. cerevisiae models, in glucose minimal
media
under
aerobic
conditions.
The glucose uptake rate was set
to 10
mmol / (gDW-hr). The apparent enhancement for E. coli inside S. cerevisiae is an artefact, caused by a short-circuit
in the proton gradient across the E.
coli cell membrane, allowing for ATP to be regenerated from ADP
at no cost.
Apparently the growth rate can be
enhanced by up to 60% by inserting E.
coli into the yeast
cytosol, compared to a maximum of the order 10% if both organisms
simply share the extracellular space. However this is an artefact caused by an unanticipated
short-circuit in the proton gradient across the E. coli cell membrane, allowing ATP
to be regenerated from ADP through the E. coli ATP synthase reaction at no
cost. This ATP can be used in both compartments (not because
there is an ATP transport reaction as such, rather there are
unanticipated couplings that effectively allow ATP to pass freely
between compartments) thus the growth-associated maintenance reactions
in both compartments can be trivially satisfied, freeing up resources
to go towards production of biomass, and artificially inflating the
growth rate.
To prove this, if we shut down all the exchange reactions the flux
through the ATPM
reaction in the yeast
compartment is still unbounded :
% perl singleopt.pl new_model.xml --close-exchanges --target=ATPM
singleopt.pl: flux unbounded, when maximising ATPM
Infinity
(The same is true for the ATPM reaction in the E. coli compartment.) However the
maximum ATPM flux is zero if we additionally shut down the ATP synthase
reaction across the E. coli
cell membrane,
% perl inspect.pl Ec_in_Sc.xml --reactions | grep ATPS4r
ATPS4r__ecoli
ATP_synthase__four_protons_for_one_ATP_ adp[ecoli_c]
+ pi[ecoli_c] + (4) h[c] <==> atp[ecoli_c] + (3) h[ecoli_c] +
h2o[ecoli_c]
% perl singleopt.pl Ec_in_Sc.xml --close-exchanges ATPS4r__ecoli
--target=ATPM
0
It is much more difficult to find a subset of reactions which are
responsible for protons leaking across the E. coli cell membrane - this is a
research exercise left for the reader! The fact that an artefact
can so easily be generated by joining models illustrates the great care
that is needed in the interpretation of the results.
The steps to put iJR904 inside iND750, both
with and without a joint biomass reaction, are included in the
Makefile for convenience (x =
0.12345 is set in the Makefile but can be changed). They can be
run by typing 'make ec_in_sc'.
Below is an
map of this model illustrated in Cytoscape, obtained using the --cytoscape
option of export.pl.
Cytoscape map of the joint model of E.
coli inside S. cerevisiae
(3752 nodes and 4161 edges). Nodes correspond to both reactions
and metabolites but only nodes with fewer than 10 links
are shown.
Portions of this site are
copyright © 2008-12 Unilever
UK Central
Resources Ltd
Registered in England & Wales, Company No 29140; Registered Office:
Unilever House, Blackfriars, London, EC4P 4BQ, UK.,
